MOLEonline web interface provides a direct access to MOLE functionality and enables on-line and easy-to-use interactive channel analysis.
MOLE 2.5 is an universal toolkit for rapid and fully automated location and characterization of channels, tunnels and pores in (bio)macromolecular structures, e.g., proteins, RNA, DNA and biomacromolecular assemblies.
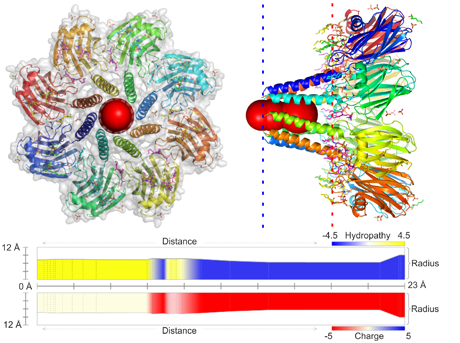
MOLEonline features
- Quickest channel calculation on the market
- Automatic transmembrane pore identification
- Layered channel profile - geometry, length and radius
- List of residues lining channels (distinguishing sidechain/mainchain contact with the channel)
- Layered or channel-wise physico-chemical properties (several types of channel radius, length, charge, polarity, hydropathy, hydrophobicity, mutability, etc.)
- Interactive selection of start or end points via XYZ coordinates, residues, or surface tetrahedron or via PatternQuery language
- Automatic active site identification using CSA for enzymes, cofactors, and cavity interiors (origins)
- Interactive visualization of structure and channels with LiteMol
- Optimization and visualization of start and end points
- Cavity and molecular surface visualization
- Download results - complete results as ZIP or PDF report, channel PDB files and scripts for PyMol, Chimera, or VMD visualization, rich JSON, and channel profile image as SVG or PNG
- Two-ways connectivity with ChannelsDB (comparison and annotation)
- Free of charge
Quick start
News
Attention
8. 6. 2019Update to Chrome 75 disables WebGL functionalities important for MOLEonline LiteMol visualization. We are working on fix on this issue. Meanwhile use other browsers for accessing MOLEonline.
Server room maintenance
17. 12. 2018We would like to announce that MOLEonline will not be available on Monday (December 17th) due to web server room maintenance. Sorry for the inconvenience.
MOLEonline update 2018 was published
1. 5. 2018Our paper, which describes the new features of MOLEonline, is accepted for publication in Web issue of Nucleic Acids Research.
New features and updates
2. 4. 2018
New features in MOLEonline:
- new Help button will allow to send us your troubled session so we can help you more thoroughly,
- multiple layers can be selected/deselected to show average properties and list of residues only around important area of the channel.
New features in MOLEonline
22. 1. 2018-
New properties
- Lipophilicity (logP and logD-like scales)
- Water solubility (logS)
- Pore mode improvements
- Better CSV export
- Residue annotation via API from Uniprot
ChannelsDB was published
1. 1. 2018The ChannelsDB database contains information about channel positions, geometry and physicochemical properties of reviewed and annotated
channels from PDB database. It was published in Database issue of Nucleic Acids Research. You can reach ChannelsDB data available for queried
PDB structure in Submission tab in right lower corner of MOLEonline GUI window.
Previous version
19. 12. 2017Old version of MOLEonline 2.0 is still available at address: http://old.mole.upol.cz/
MOLEonline is supported by ELIXIR CZ research infrastructure project (MEYS Grant No: LM2015047)
Sponsored by ELIXIR-CZ, UPOL, MUNI




