Walkthrough guide
Loading structure
You can load the structure of biomacromolecules according to the PDB ID code (e.g. 1TQN) and structure will be automatically loaded from Protein Data Bank in Europe using CoordinateServer service (http://www.ebi.ac.uk/pdbe/coordinates/). Potentially, you can also add the biological assembly ID specification (e.g. 1, 2, PAU, etc.).
MOLEonline can also process user-submitted files with your manually refined structure as a PDB or mmCIF file. In this case you can use also the GZIP archives up to the maximal size of the file 100 MB, for example you can prepare structure only with chain A with removed ligand molecule, however such behaviour can be mimicked by the service settings.
How to choose starting point
Firstly you can choose from the automatic starting points:
- CSA – residues from the Catalytic Site Atlas (http://www.ebi.ac.uk/thornton-srv/databases/CSA/)
- Origins – either CSA Origins (green balls) or Computed origins in each cavity (blue balls)
- Cofactor starting points
Or use your own starting point:
- Exact residue, atom or point – you can choose it directly in the structure by clicking to the position
- Choose residue in the Protein Sequence
- Choose the tetrahedron on the surface of the structure by Ctrl+left mouse click in case you intend to compute pores or paths.
Submit the calculation
- Submit button is situated in the bottom left corner
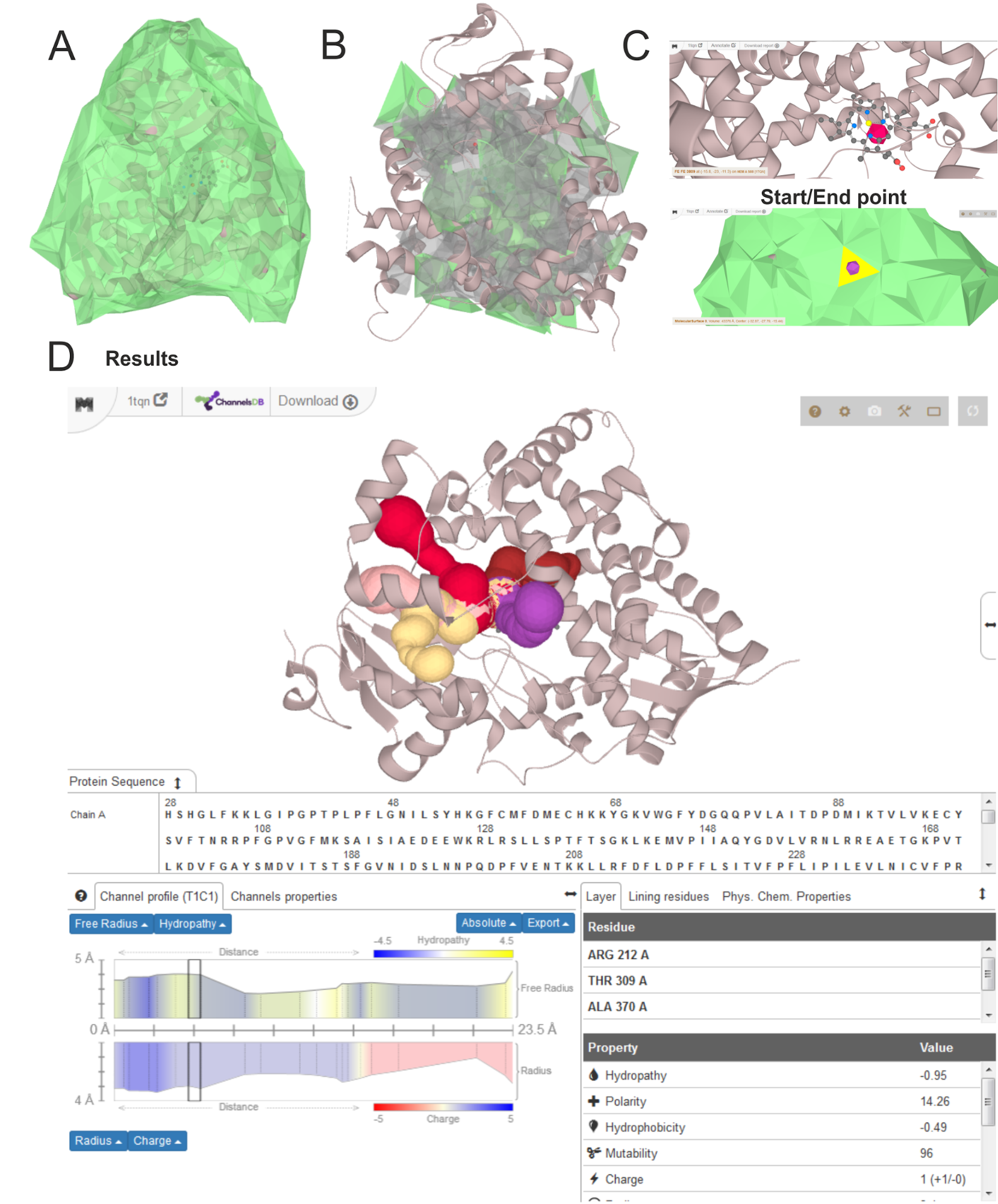
A - clickable surface defined is shown in green; B - detected cavities with clickable surface in green, C - search for the starting point from residue or clickable facet with use of Control key; D - resulting channels with properties of selected second shortest channel (T1) leading from the largest cavity (C1).
Visualization
MOLEonline uses LiteMol Viewer for the visualization of the structure and the computed channels or pores. The LiteMol provides the intuitive tool for the easy manipulation with the visualizing structure. Moreover, LiteMol allows using several types of representations such as sticks, cartoon, lines etc.
Download the results
You can download the report of the computed channels or pores by clicking Download Report button on the upper left corner of the visualization window in several formats. You can choose from: Molecule, PyMol, VMD, PDB, Chimera, JSON or the whole report (all computed data including structures, channel profiles and physicochemical properties) in zip format.
You can also download the profile of the individual channel by clicking to the Export button next the profile and choose if you want to obtain the PNG or SVG.